Just joining us? Start with part one of our demystifying synthetic biology series.
Synthetic biology seems like a brand-new, precision scientific field – just bursting with potential – but is it really all that new? Biology is often seen as the unpredictable cousin of the math-based sciences chemistry and physics. But it’s exactly this inherent variation within biological systems that makes life uniquely adaptive and resilient – though it can also make these systems painfully difficult to understand. Only in the past century have researchers really understood the significance of DNA – the hard drive within all living cells that stores the information for life. Molecular biology research of the 20th Century has developed from crude tools into precision techniques which, when combined with new technologies in reading and writing DNA, has resulted in the field of synthetic biology.
We now have the ability to make accurate predictions on an organism’s physiology and metabolism just from looking at its genetic and product profiles, allowing us to manipulate or engineer those genes and pathways more rationally toward practical applications. Contrary to popular belief, synthetic biology is not a wholly new field. To paraphrase Sir Isaac Newton: if synthetic biology sees further, it is because we stand on the shoulders of molecular biology giants. This field stands atop the shoulders of technological titans in biology, chemistry, physics, engineering, and computer science. As part of our continuing blog series on demystifying synthetic biology, we will explore the basic approach to engineering an organism, the common research tools used in the field, and how they have advanced to enable synthetic biology.
Reading and Writing in the Language of Life
Cells are a lot like living factories, and at their core, the instructions are written in the universal language of life: DNA. DNA acts like a computer storing files for programs on a hard drive, but instead of 1s and 0s, DNA uses four nucleic acids – A, G, C, and T – which are arranged into genes that encode proteins. Collectively, all the genes from an organism make up its genome, and all the proteins make up the proteome. These proteins fill various roles, including forming the basic cellular functions like metabolic pathways.
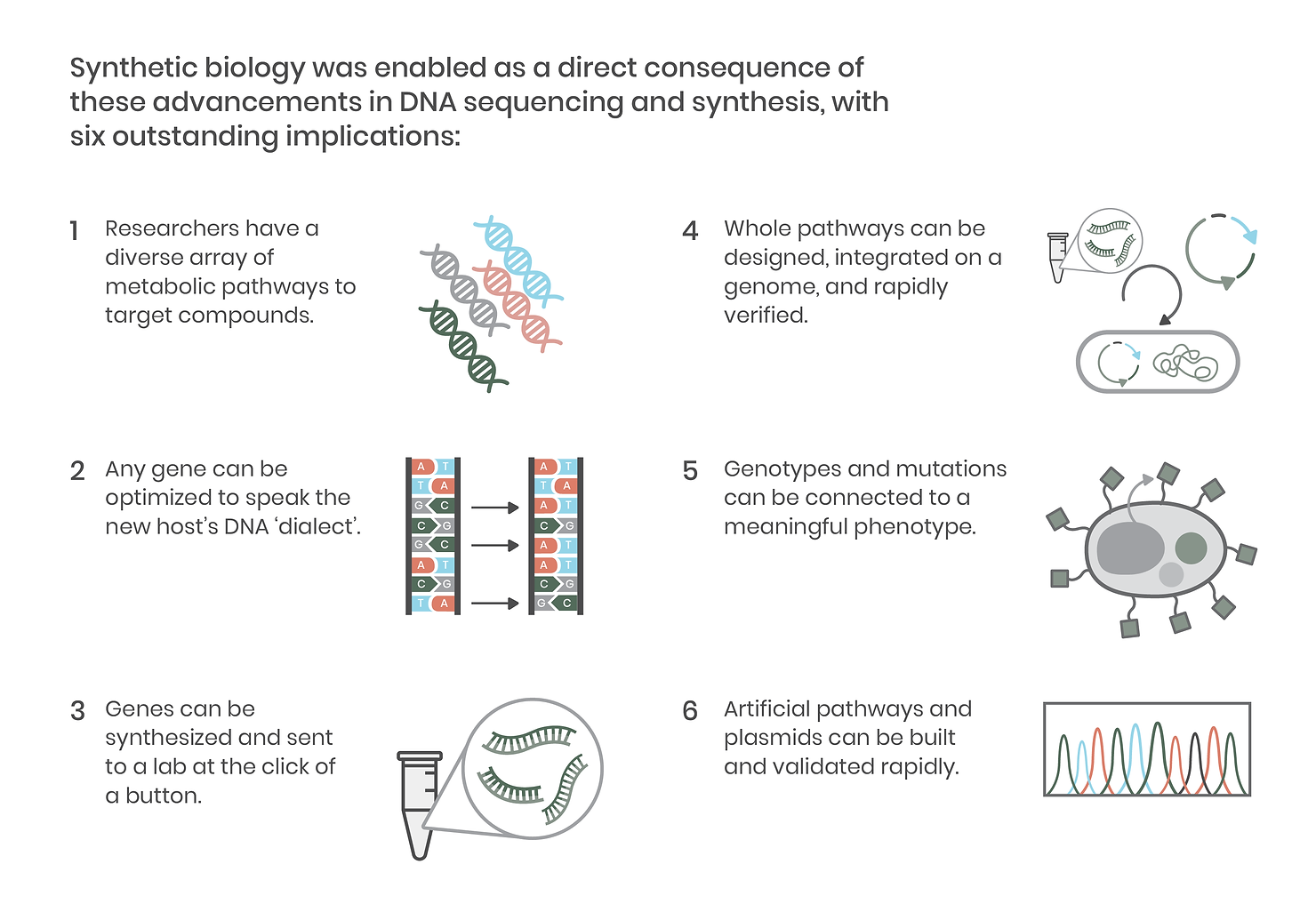
Reading DNA – sequencing the precise AGCTs that make up a strand of DNA like a gene – is a critical element of synthetic biology. The original Sanger sequencing method took a day to read just 1,000 nucleic acid letters. Fortunately, next-generation sequencing (NGS) rapidly improved throughput, and with today’s methods, we can read whole genomes in a single day for a fraction of the cost. This development made genes (and genomes) of diverse organisms accessible to laboratories around the world, revealing new metabolic pathways creating compounds of interest across thousands of plants, animals, microbes, and viruses. It also meant that labs could more precisely make and validate DNA constructs, leading to the development of more accurate, high-throughput molecular biology techniques and paving the way for advanced genetic engineering tools like CRISPR.
Equally as critical as DNA sequencing is the ability to synthesize it, block by block. DNA synthesis has markedly improved accessibility to the diverse genomes revealed by the NGS sequencing mentioned above. On a practical level, this means researchers no longer have to grow an organism or its cells to obtain a genetic template. DNA synthesis also facilitates the rapid construction of metabolic pathways that can be assembled onto a single plasmid vector and introduced into an organism.
Since the development of improved sequencing technologies and the increasing accessibility of DNA synthesis, integrating genes and whole pathways into organisms has become commonplace in biological research. Together, these technological advancements have enabled precision molecular biology and, in doing so, ushered in the era of synthetic biology.
Biofactories in Nature
Going back to our factory analogy, proteins – specifically, enzymes – act as the factory workers in a cell. They use sugar, like glucose, or other compounds as raw materials to generate metabolic byproducts called metabolites. In a metabolic pathway, there may be several intermediate metabolites generated as different enzymes work together, similar to an assembly line. Enzyme A converts sugar into Compound 1, Enzyme B then turns Compound 1 into Compound 2, and so on until the final metabolite is reached. Humans have taken advantage of these natural processes industrially, such as in brewing beer. Beer brewing is a metabolic pathway in action. Yeast turns sugar into alcohol via a series of enzymes under the right growth conditions for fermentation.
The natural pathways of some plants and microbes generate intermediate metabolites that are precursors for other beneficial molecules. In synthetic biology, we take advantage of widely available data such as genome sequences and the metabolites those organisms produce. From there, we can determine which genes encode the enzymes for specific metabolic pathways. By adding genes from different organisms, we can redirect precursor molecules from the host organism into an entirely new pathway, producing novel and useful compounds. Through synthetic biology, making something entirely new becomes as straightforward as brewing beer. Of course, to achieve this, we must have tools that allow us to precisely harness the power of nature.
Harnessing the Power of Nature with Synthetic Biology
We complain that biology is often unpredictable but, until recently, the tools we used to study it have been clumsy and crude compared to the precision tools and techniques we employ in the field today. Not long ago, molecular biology was a messy science fraught with complications and potential error. For example, if you wanted a specific gene, you had to grow the organism first to extract its DNA. You would then use PCR (like a copy machine) to amplify a fragment and then use restriction enzymes (DNA cutters) to snip the ends of the DNA before pasting it into a target destination, like a plasmid vector for expression in a new host.
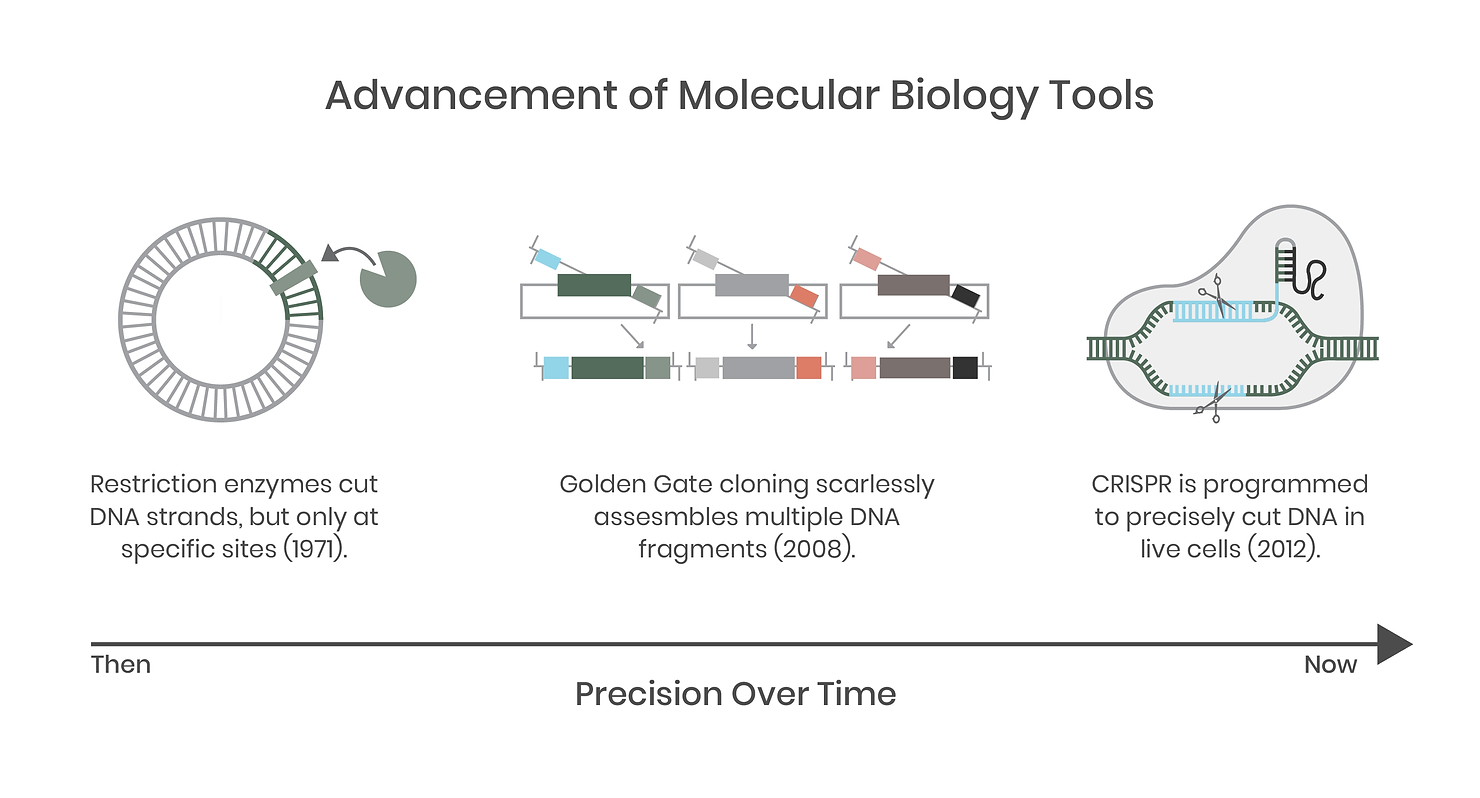
In synthetic biology today, many of these tools have improved markedly and are still in use, but are backed up by DNA synthesis, standardized genetic parts (genetic regulatory elements such as promoters and terminators), modular systems such as Golden Gate assembly, and gene editing systems like CRISPR. Genomic integration of DNA is now commonplace compared to 25 years ago. This is enabled by technologies such as CRISPR, a specific and targeted DNA-cutting molecular tool that allows precise genetic engineering of previously intractable host organisms. Ultimately, these tools have improved the speed, fidelity, and complexity of molecular biology, which has evolved to enable an engineering approach to biology and underpins the entire field of synthetic biology.
Biology is fundamentally about understanding how life, nature, and our ecosystem work. Mother Nature did pretty well before humans came along. If we can harness the power of nature through synthetic biology – specifically the precision editing of organisms to solve problems – we may be able to undo some of the damage mankind has caused during our time, while still enjoying the benefits that we created along the way.
Next time…
Now that we’ve seen how synthetic biology is simply the next generation of molecular biology tools developed in the 20th Century, we will explore how synthetic biology makes use of other 21st Century technology to scale R&D and allow rapid innovation of bio-based solutions. The greatest strength of this field is its ability to learn from data and iterate the R&D process in a rational, engineering-like approach.
In our next blog of this series, we will discuss how automation and software are applied in synthetic biology, how these technologies help scale the molecular biology tools underpinning the field, and where their values and limitations lie.
Follow Antheia on LinkedIn to stay up to date on our newest demystifying synthetic biology blogs.